DNA - An overviewPage
11
11
Slide 68
When an unpaired or incorrectly paired base are clip off by exonucleases.
When an appropriate base-paired terminus results, polymerase begins resynthesis by adding nucleotides to the 3’ end.
The 5’ to 3’ exonuclease activity of many prokaryotic DNA polymerases is also very important.
It functions in the removal of segments of DNA damaged by UV and other agents.
Slide 69
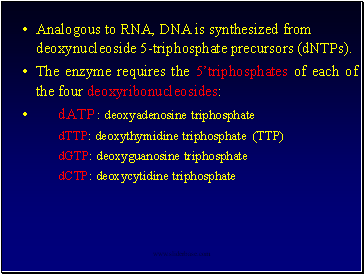
Analogous to RNA, DNA is synthesized from deoxynucleoside 5-triphosphate precursors (dNTPs).
The enzyme requires the 5’triphosphates of each of the four deoxyribonucleosides:
dATP : deoxyadenosine triphosphate
dTTP: deoxythymidine triphosphate (TTP)
dGTP: deoxyguanosine triphosphate
dCTP: deoxycytidine triphosphate
Slide 70
This enzyme is active only in the presence of Mg+ ions and preexisting DNA.
This DNA must provide two essential components, one serving a primer function and other a template function.
1. Primer DNA: DNA polymerase I cannot initiate the synthesis of de novo. It has an absolute requirement for a free 3’hydroxyl on preexisting DNA chain.
DNA Polymerase I catalyzes the formation of a phosphodiester bridge between the 3’OH at the end of the primer DNA chain and 5’phosphate of the incoming deoxyribonucelotide.
The direction of synthesis is always 5’ to 3’
2. Template provides ssDNA that will direct the addition of each complementary deoxynuceotide
Slide 71
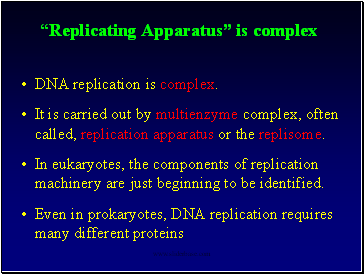
“Replicating Apparatus” is complex
DNA replication is complex.
It is carried out by multienzyme complex, often called, replication apparatus or the replisome.
In eukaryotes, the components of replication machinery are just beginning to be identified.
Even in prokaryotes, DNA replication requires many different proteins
Slide 72
Replication fork: The junction between the newly separated strands and unreplicated double stranded DNA
Leading and Lagging strand: Due to the anti-parallel nature of DNA, one strand will synthesis continuously towards replication fork and other strand will synthesis discontinuously away from the replication fork.
The continuously synthesizing strand is called leading strand and discontinuously synthesizing strand is called lagging strand.
Okazaki fragment: A short fragment of DNA formed on the lagging strand during replication is called Okazagi fragment. It will be around 100 – 1000 bp in length. In eukaryotes it identified about 100-200 nucleotides length.
Processivity: The ability of an enzyme to catalyze many reactions before releasing its substrate is called processsivity
Contents
- Gene
- What are the requirements to fulfill as a genetic material?
- The Genetic material
- Griffith experiment
- Rough
- The Hershey – Chase Experiment
- RNA as genetic material in small viruses
- Dna Structure
- Chargaff’s rule
- X ray diffraction
- Double Helix
- Semiconservative Replication of DNA
- The Meselson – Stahl Experiment
- Models of dna replication meselson and stahl expt.
- Cairn’s Experiment
- John Cairns
- Unique origin and Bidirectional replication
- The Replication of DNA
- Dna Polymerases
Last added presentations
- Space Radiation
- Practical Applications of Solar Energy
- Solar Thermal Energy
- Newton's Laws
- Newton’s third law of motion
- Resource Acquisition and Transport in Vascular Plants
- Gravitation